A New Model for DNA Damage in C. elegans Breaks Down Radiation Effects
By XU Zhao
Recently, a research team led by associate Prof. XU Zhao from the The Institute of Nuclear Energy Safety Technology, Hefei Institutes of Physical Science, Chinese Academy of Sciences (INEST) successfully constructed a nanodosimetric model of C. elegans' germ cells, quantifying photon-induced DNA damage and proposing a dose-dependent DNA damage repair model based on target theory.
The relevant results were published in Radiation Physics and Chemistry.
Energy deposition at the subcellular level is crucial in understanding radiation's biological effects. However, current dosimetry methods don't fully capture these effects. To improve this, it's important to study the track structure of particles at the nanoscale, particularly how radiation damages DNA. C. elegans is a key model for radiation studies, as its germ cell damage is similar to human DNA damage, though DNA models for its germ cells are still lacking.
In this study, researchers focused on germ cells and used the Hilbert curve method to construct a DNA model, incorporating various parameters such as physical constructional function, energy threshold models, and free radical scavenging distances to analyze photon-induced DNA damage. After testing different configurations, they selected constructor function 4, a linear energy threshold model, and a 9 nm free radical scavenging distance as the optimal simulation parameters for photon exposure. Using GEANT4, a software designed to simulate particle interactions with matter, the researchers quantified the generation of Double Strand Breaks (DSBs). At an irradiation dose of 10 Gy from 137Cs, they observed an average of 31.6±2.2 DSBs.
Additionally, the team compared the DSBs induced by 137Cs radiation at different doses to experimental data and proposed a dose-dependent DNA damage repair model for nematode germ cells based on target theory. They successfully fitted the total DSB damage and DSB cluster damage repair processes, and the results closely matched existing biological experimental data.
This work provides an important means for understanding the biological effects induced by ionizing radiation.
Fig. 1. The illustration of nucleus of C.elegans.
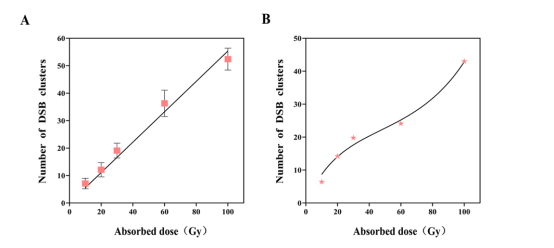
Fig. 2. Fitting the number of DSB cluster damages in a germ cell of C.elegans. (A) DSB original damages, (B) DSB damages after repairing.














